Deep Learning Reconstruction of Optoacoustic images on FPGA
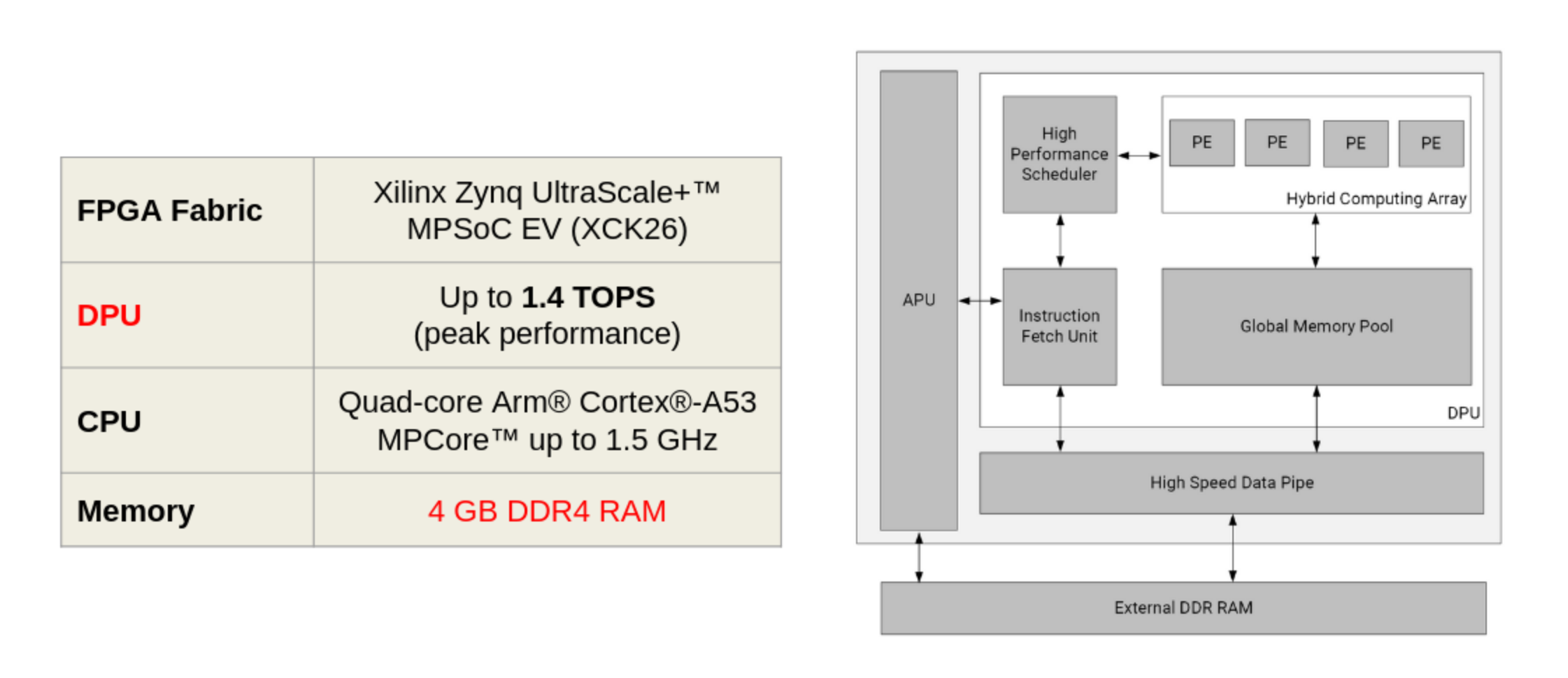
Deep Learning on AMD Kria™ K26 SOM optimized for the DPU
Overview
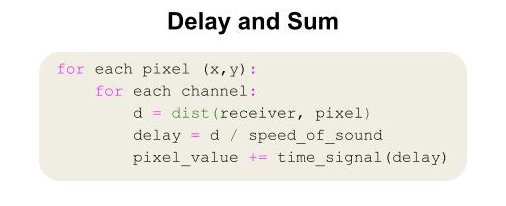
The standard reconstruction of optoacoustic images (similar to ultrasound) is Delay and Sum, an inverse problem that requires high computational load for high-resolution imaging. While advanced imaging became possible over the last 20 years thanks to increased computational power, Delay & Sum is surprisingly inefficient on GPUs. This stems from non-trivial indexing and irregular memory access patterns caused by the data dependencies involved in this method—each pixel requires signals from many sensors.
My goal is to develop a real-time image reconstruction using deep learning, optimized for today’s AI accelerators.
Optoacoustic
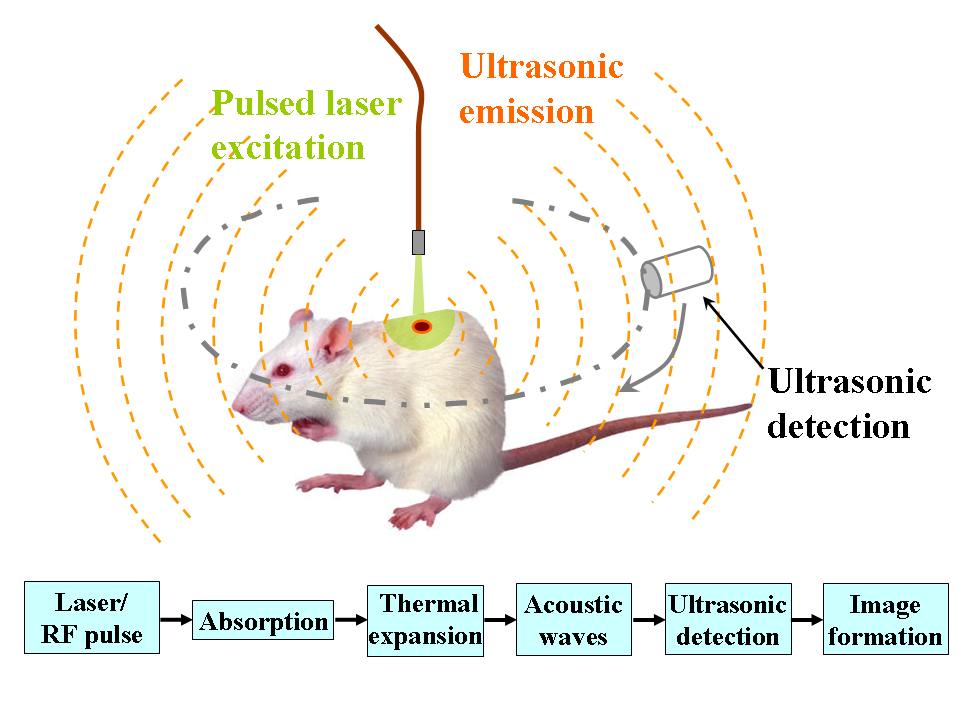
Optoacoustic imaging is similar to ultrasound, but instead of sending and receiving a pressure wave, we create one by focusing a laser pulse at a specific point. This causes thermal expansion, generating a pressure wave that travels back to be received by piezoelectric sensors.
This imaging modality is non-ionizing, making it completely safe, and can offer high resolution at both the vascular and molecular level—enabling applications like measuring oxygen levels and detecting early-stage cancer.
Dataset
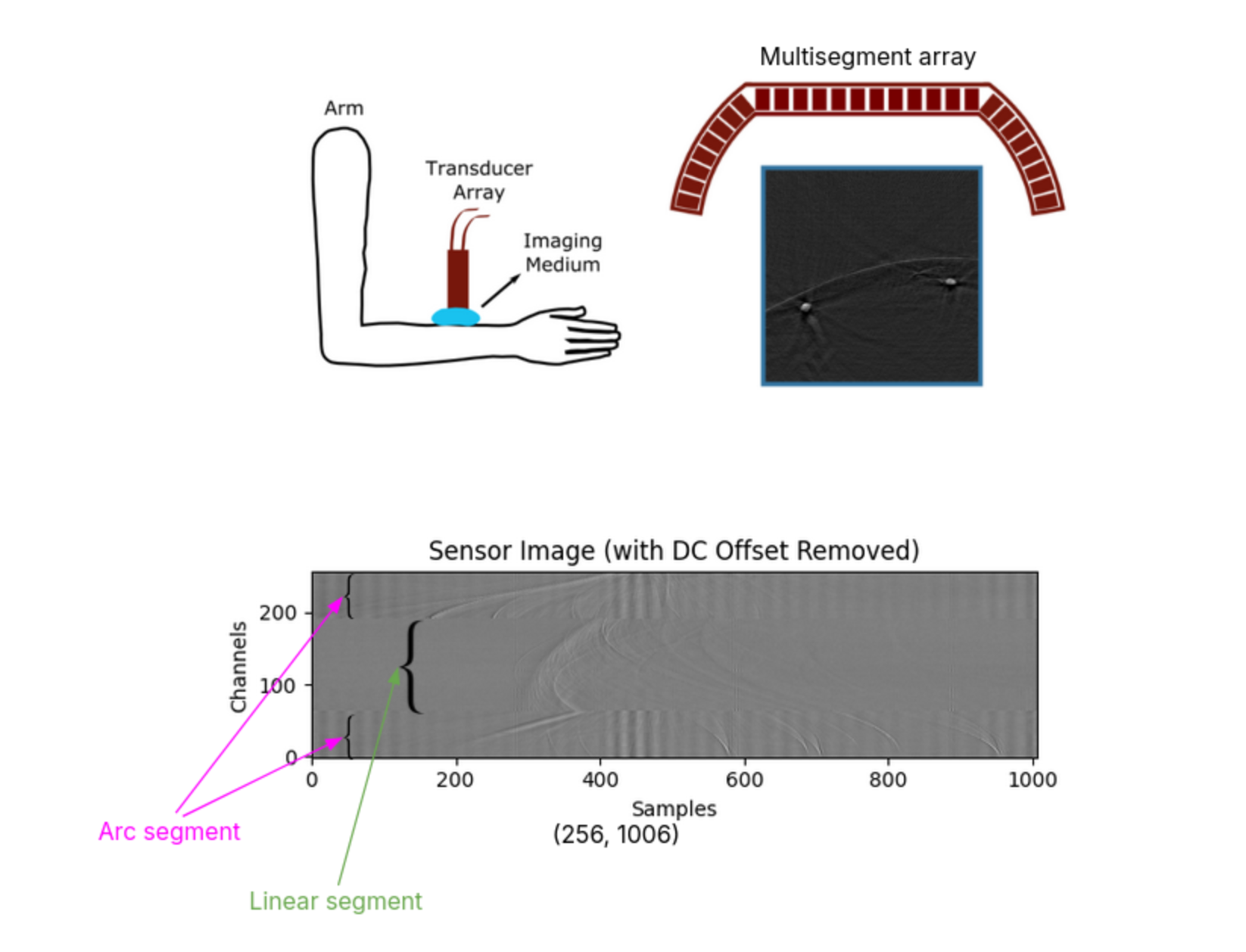
The data was collected at Professor Daniel Razansky’s Lab with recordings from human right and left arms. In total, I worked with around 5,500 float32 images. They were recorded using a multisegment array—as shown in the figure below—with a linear section and two arch sections, totaling 256 channels.
Target Device
I used the Kria K260 Robotics Kit to deploy the model and run the inference. This is a System On Module (SOM) from AMD dedicated to high-performance and intelligent industrial applications. Its most attractive feature is the Deep Learning Processing Unit (DPU), which enables real-time imaging. The DPU is essentially a microcoded compute engine with an optimized instruction set for machine learning operations, especially convolutions.
Model Design and Deployment
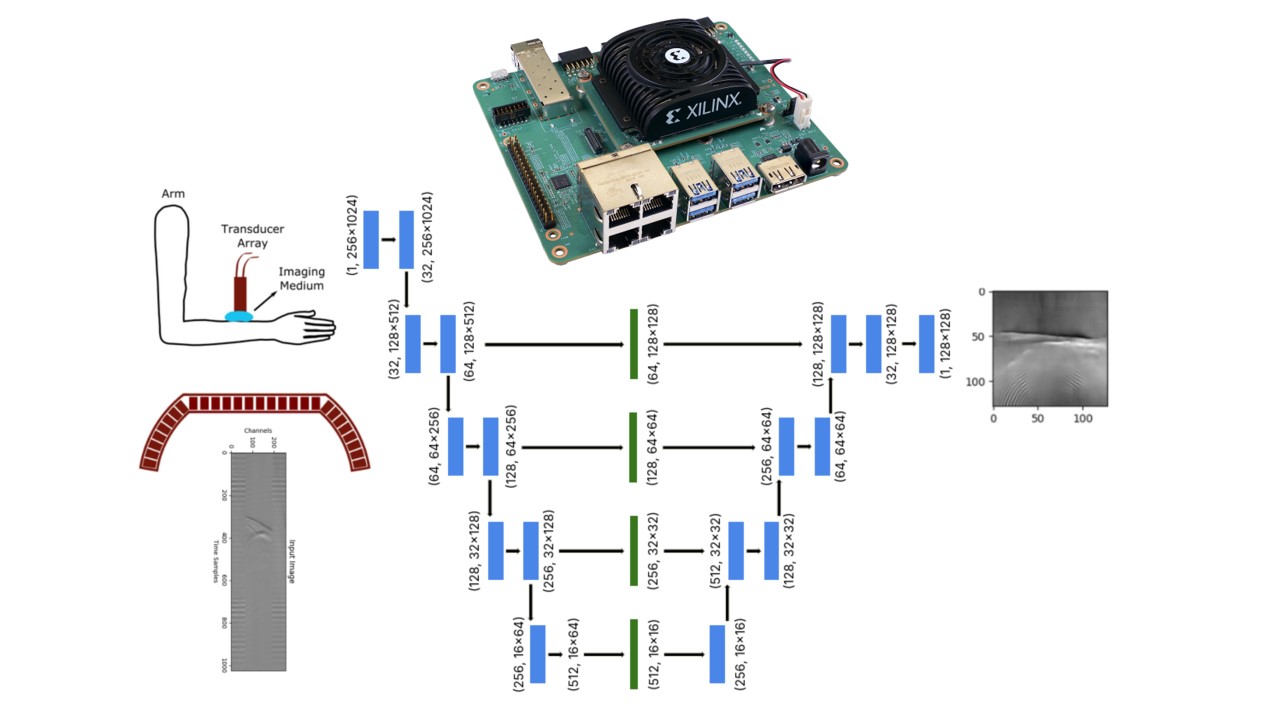
- Model Selection: Currently, the best network architectures for biomedical image segmentation and reconstruction are U-Net autoencoders. I chose this because it’s fast, precise, and CNN-based, which is optimal for the DPU.
- Preprocessing: First, to generate my ground truth images, I used standard delay and sum and normalized the images. For my time series pressure signal input, I started with DC offset removal, then applied median padding to resize images from (256×1006) to (256×1024) for power-of-2 compatibility, followed by normalizing to zero mean and unit variance, and finally removing outliers caused by bad recordings.
- Hardware Adaptation: The key challenge here was ensuring all inference operations ran on the DPU rather than being offloaded to the CPU. This meant using only DPU-supported operations—specific kernel sizes, no adaptive pooling, no resizing, no custom functions, etc.
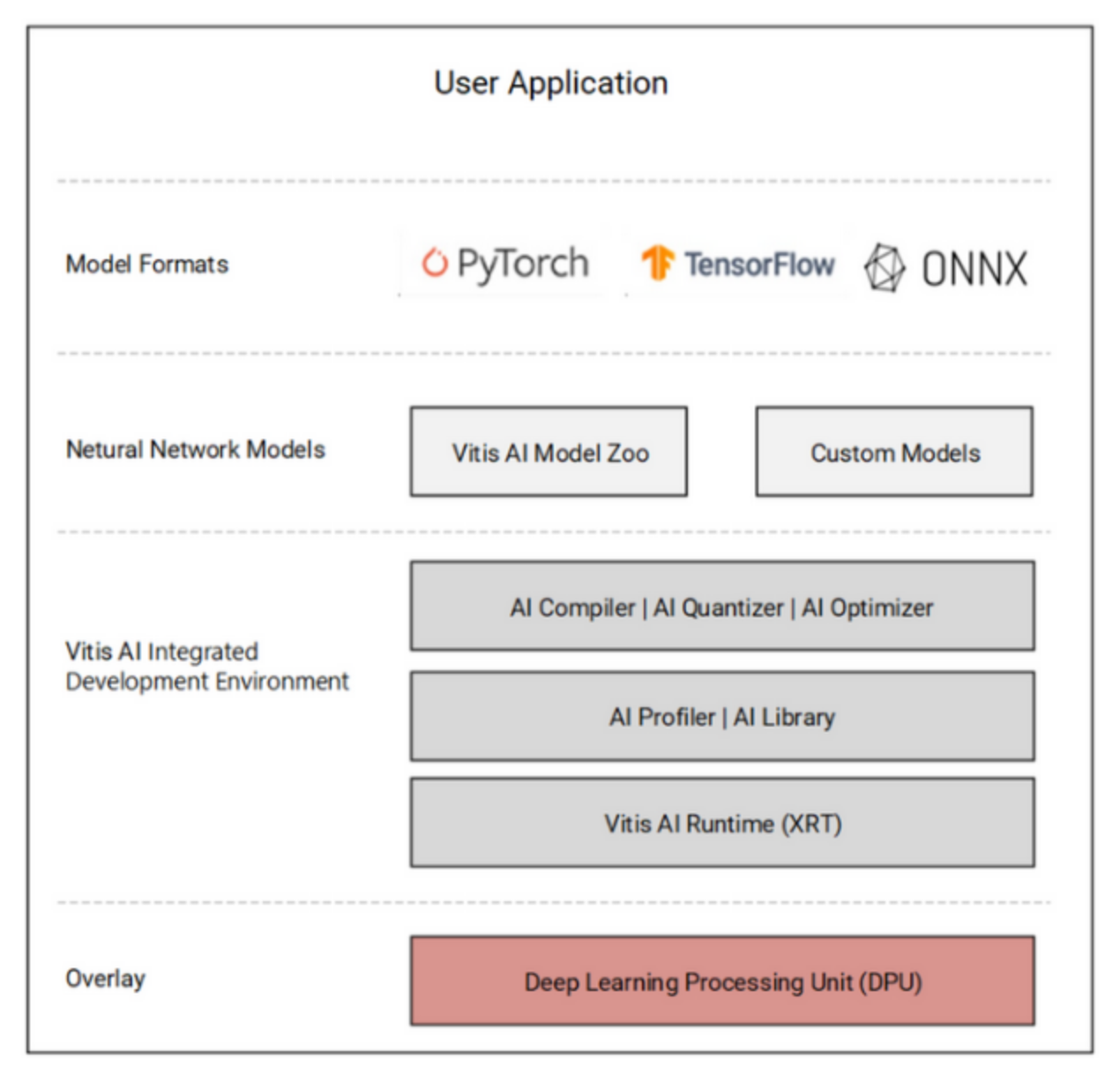
- Training and Deployment: Training was done offline using PyTorch. The final model was then transformed into ONNX format and quantized to INT8 using Vitis AI, then compiled to create a deployable .xmodel file, which is essentially a mapping of the neural network to the specific hardware architecture. Vitis AI is the IDE for deploying pretrained models quickly on AMD platforms and accelerating inference on the DPU without working on bare metal.

Results
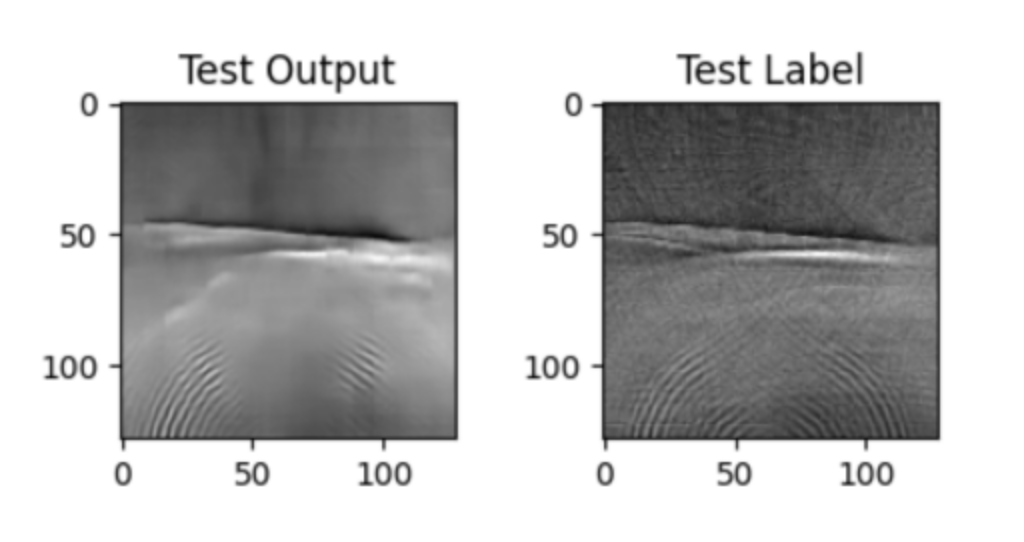
As shown in the figure above, the reconstruction on the test set successfully exposed the main features of the image. We can see the main reflecting line in the middle and the arches on the bottom of the picture. The model evaluation resulted in:
- Mean Square Error (MSE): 0.567
- Parameter Size: 7.5 MB (much smaller than the 4 GB DDR4 memory)
- Inference time: due to time constraints, I didn’t evaluate it on the Kria; however, on my laptop I was able to reach 6 fps, which is already promising—it will definitely achieve real-time performance on the DPU.
Finally, I also explored sub-channel sampling using only the signal from the linear segment for reconstruction, which resulted in a smaller model (about half the size) but naturally slightly worse image quality with an MSE of 0.7.